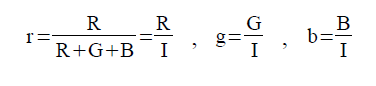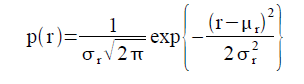Video and Audio
Video and audio refer to storage formats for moving pictures and sound, respectively, which changes through time. Recording a video or audio is also known as video or audio codecs. Video codecs comprises of a series of images, or frames while audio codecs commonly comprises of a single channel or mono, two channels or stereo or more. The quality of videos depends upon the number of frames per second, the resolution of the images and the color space used. On the other hand, the number of bits per playback time, or bitrate, determines the quality of audio [1].
Video and audio are used to develop family videos, presentations, web pages and others. It is recommended by the Web content accessibility guidelines to provide alternatives for this kind of media like captions, descriptions or sign language when producing videos all the time [1].
Video and audio are formulated to improve experiential learning or entertainment.
Video is a series of still images presented in a fast succession so that there will be a perception of motion by the audience [2]. Observe the image below.
Figure 1. A GIF image of a dog [3].
A video can either be analog or digital. In this regard, the image processing techniques we have learned can be applied to the ‘still’ images. The frame rate for a digital is known as fps or frames per second. By taking the inverse of fps, we get the time interval between frames or the Δt [2].
This activity revolves around basic processing of a video. The audio here is omitted.
In particular, dynamics and kinematics of a specific system will be extracted from a video [2].
Important note: If an image seems to be indistinct, simply click that image for larger view. 😀
Kinematic Experiment
A video of a kinematic experiment, specifically 3D Spring Pendulum, is the subject for this activity.
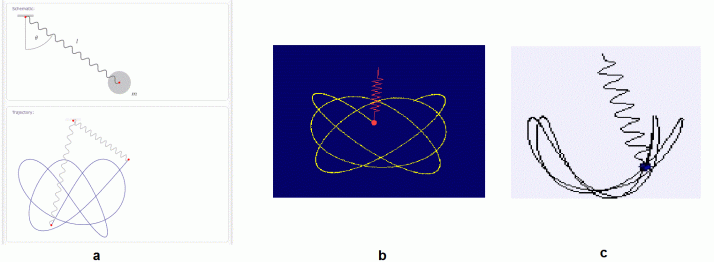
A 3D spring pendulum consists of a spring-mass system with three degrees of freedom. Having such, its behavior becomes chaotic. For a simple pendulum, the string length is constant giving rise to a constant period. However, the length of spring in a spring pendulum changes every time [4]. Below are some examples of chaotic behavior.
Figure 2. Chaotic behavior of two 3D spring pendulum systems. a from [4], b from [5] and c from [6].
Materials and Setup
The materials used are iron rod, clamp, spring, 20 g mass (actually 19.1g), Canon D10 camera, tripod, FFmpeg software, laptop, red pentel pen and masking tape. The assemblage of the first four materials is shown below.
Figure 3. 3D spring pendulum (drawn using MS Powerpoint).
The digital camera was used to take the video of the kinematics and the software for video processing. The red pentel pen and masking tape were used to ensure that the color of the mass is unmistakably different from the background.
Procedure
The first step is to take a video of the actual 3D spring pendulum system in motion. The video showing the system along the side of the setup can be found here:
http://www.mediafire.com/download.php?6xfzap395c99w6z
The frame rate of the video is actually 30 fps. FFmpeg was used to extract images from the video above. This program was ran using command prompt and following the format:
ffmpeg -i <filenameofimage> <filenameofsavedimage>
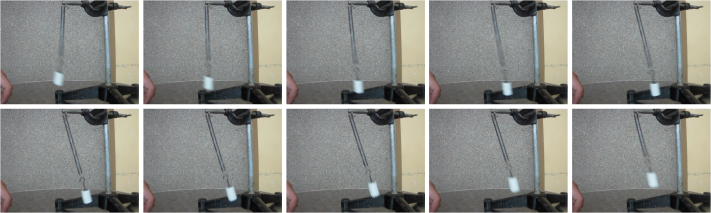
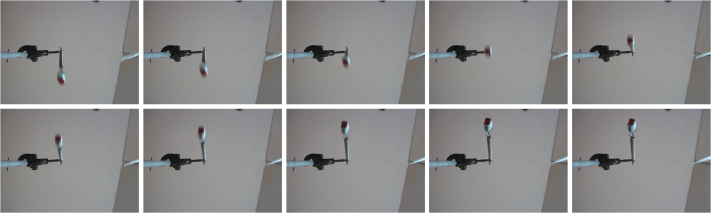
The 141st to 150th frames/images from the video are shown below.
Figure 4. Sample images extracted from the video above (141st to 150th frames).
Since the images were now extracted, image processing techniques can be executed. Here is my plan to get the position of the mass:
- loop through images
- image segmentation (parametric and non-parametric then choose the better one)
- get the pixel position of the centroid of the blob
- append the pixel positions to an array and plot the track of the mass in 2 dimensions
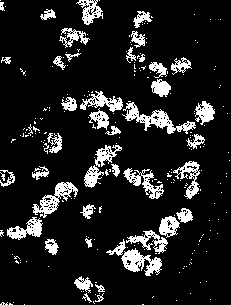
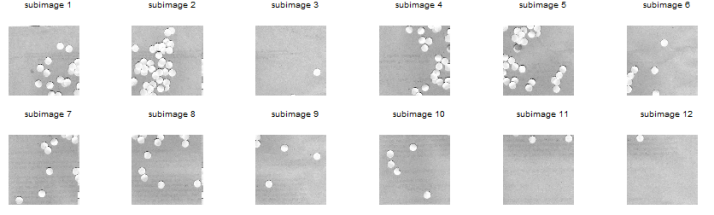
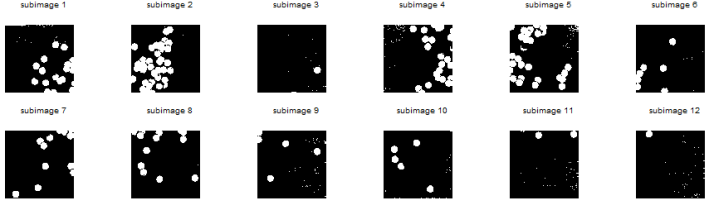
So the next thing I did was to do Image Segmentation. But here’s the problem: I found out that the iron rod was also included in the segmentation! 😦 Since the mass was white and specular reflection occurred in the iron rod, the segmented images contain the iron rod as well. I cropped out the image so that there will be no more problems. However, some of the pebbles were also included! 😦 I used FastStone Photo Resizer 3.1 to crop and edit the images by batch. We were not careful about the color of our subject that is why we have to deal with this problem.
Figure 5. Cropped version of the image from Figure 4.
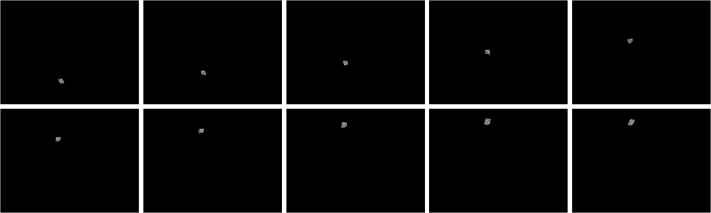
What I did was to segment the images first. The segmented version of the images from Figure 3 using parametric segmentation are shown below.
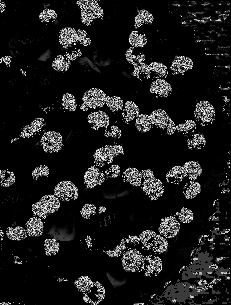
Figure 6. Parametrically segmented images corresponding to the images in Figure 5.
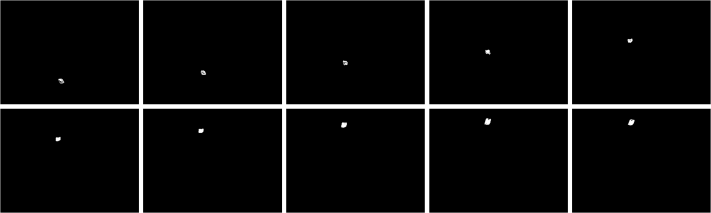
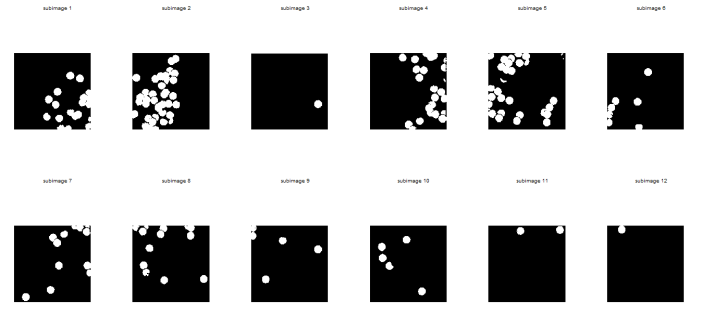
I thought of using morphological operations for the images above so that I can isolate the biggest blob. However, I still have another choice. That is, non-parametric segmentation. Figure 7 shows the corresponding segmented images using non-parametric segmentation.
Figure 7. Non-parametrically segmented images corresponding to the images in Figure 5.
By inspecting Figures 6 and 7, we can see that non-parametric segmentation produced better results so I used it for the next step. The next thing we need to do is to get the centroid of the ROI (region of interest) which is the mass so that we can know its pixel position in the image.
By the way, segmentation was done with ease because I looped through the images. 😀 The processing is simple but running the code took a very looooong time. The video’s length is 3 minutes 11 seconds but I only processed the first minute. That means I looped through 1800 images in order to produce both the parametrically and non-parameterically segmented images. 🙂
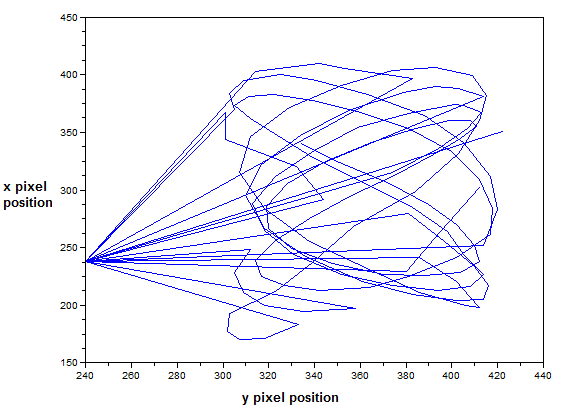
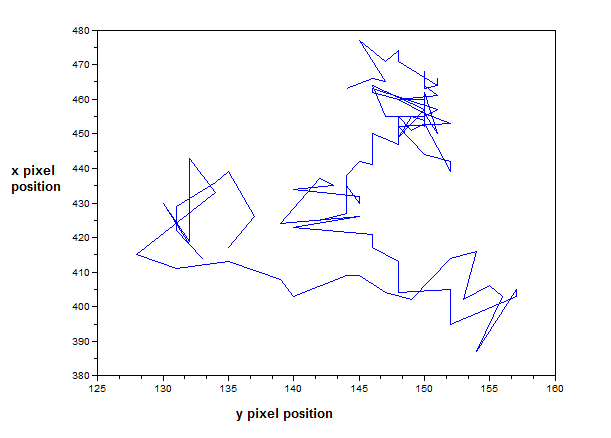
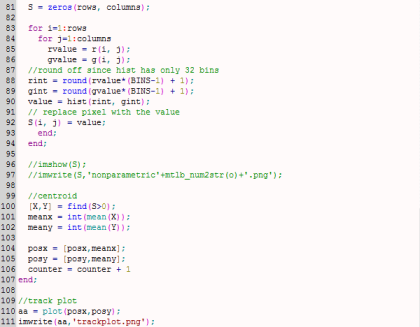
[Going back to the discussion] I made two empty arrays from the beginning of my code: posx and posy. See the last figure for my Scilab code. When the centroids of all the images were taken, the x and y positions are binned in posx and posy, respectively. In the end, posx and posy are plotted against each other. The result is actually the track plot and it is shown below.
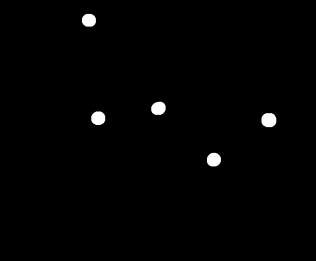
Figure 8. Track plot of the mass in the pendulum from the video above (170 frames).
The plot from Figure 8 shows the track of the mass using 170 frames only so that it will be presentable. Too much frames will show more lines that might conceal the directions. We can see from Figure 8 that the path of the mass is chaotic and similar to Figures 2a and 2b.
We took another video of the 3D spring pendulum but this time, its motion is viewed from the bottom of the setup. It can be found by clicking the following link:
http://www.mediafire.com/download.php?p86nd84r95zcjyh
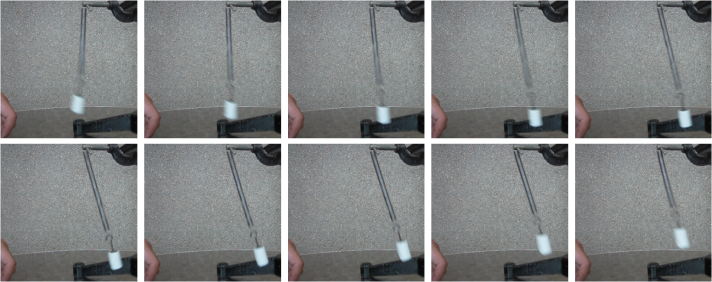
The 141st to 150th frames are shown below.
Figure 9. Sample images from the second video (141st-150th frames).
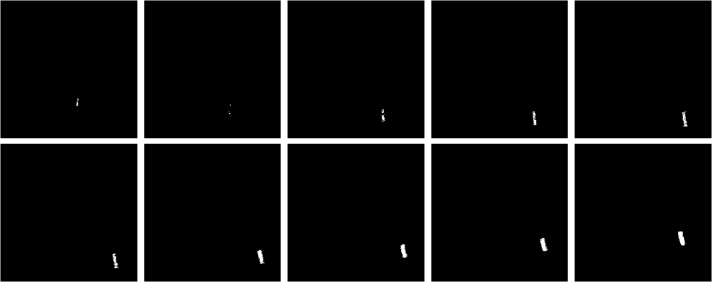
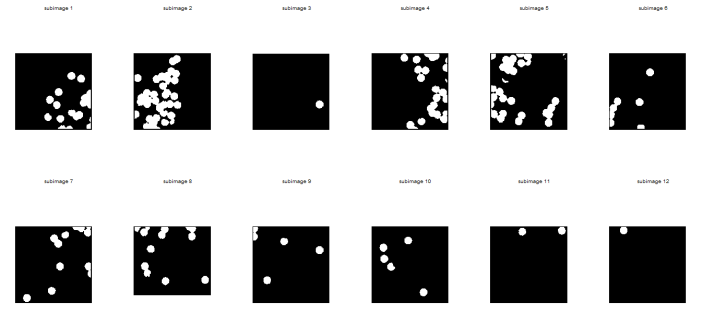
I did not have a problem segmenting the images from this scene since the mass here is covered with masking tape applied with red pentel pen ink. (Thank God.) The parametrically segmented images corresponding to the 141st to 150th frames are shown below for sample images.
Figure 10. Parametrically segmentated images corresponding to the images from Figure 9.
and the non-parametrically segmented images are shown in the next figure.
Figure 11. Non-parametrically segmented images corresponding to Figure 9.
Again, the non-parametrically segmented images were better than the parametrically segmented ones just like in the case of the first video. I then looped through the images and took their centroids to determine their positions per image. The plot of the positions are shown below.
Figure 12. Track plot of the mass in the pendulum from the second video (100 frames).
The plot from Figure 12 shows the track using 100 frames only. Adding more frames will show more chaotic plot. Once again, the track of the mass is similar to the track illustrated in Figure 2. We can account the difference from the fact that it is not ideal and by presence of air drag, spring constant, initial force and other external forces.
On a side note, I thought of using correlation from the beginning instead of taking the centroid. This method, however, consumes too much memory and running time so I chose the determination of centroid in the end.
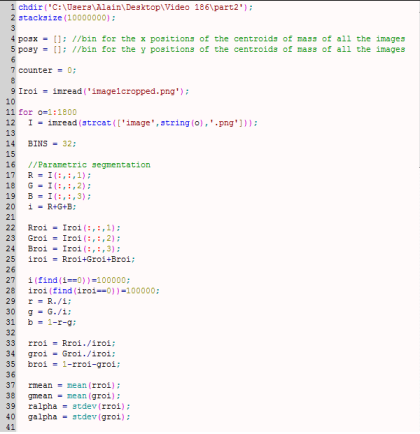
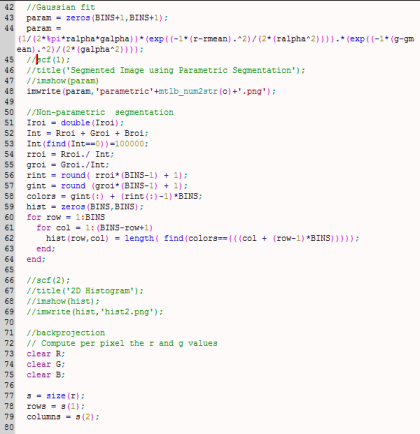
My whole code is shown below.
Thanks to my cooperative groupmates, Gino Borja and Tin Roque. We prepared the kinematics experiment and took the video together. Gino has introduced FFmpeg. We have thought of looking for the path/track of the mass together but we decided to choose our own way to attack the problem and create the programming codes on our own. We would like to thank the VIP group of IPL for lending their Canon D10 camera and a tripod. I would like to give myself a grade of 10 for doing all the steps.
This is the last activity for this course! Yey!! 😀 I can say that I really enjoyed this subject and I’ve learned a LOT about image processing and Scilab. This course also extended my imagination! 😀 Thank you!
References:
1. “Audio and Video”, retrieved from http://www.w3.org/standards/webdesign/audiovideo.html.
2. Maricor Soriano, “AP 186 Activity 12 Basic Video Processing”, 2012.
3. “Puppy/Dog Animations”, retrieved from http://longlivepuppies.com/PuppyDogPicture.a5w?vCategory=Gifs&bPostnum=00000000222.
4. “From Simple to Chaotic Pendulum Systems in Wolfram|Alpha”, retrieved from http://blog.wolframalpha.com/2011/03/03/from-simple-to-chaotic-pendulum-systems-in-wolframalpha/.
5. “Spring Pendulum”, retrieved from http://www.maths.tcd.ie/~plynch/SwingingSpring/springpendulum.html.
6. “EJs CM Lagrangian Pendulum Spring Model”, retrieved from http://www.compadre.org/osp/items/detail.cfm?ID=7357.